UBE3A
| UBE3A | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | UBE3A, ANCR, AS, E6-AP, EPVE6AP, HPVE6A, ubiquitin protein ligase E3A, PIX1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 601623 MGI: 105098 HomoloGene: 7988 GeneCards: UBE3A | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Ubiquitin-protein ligase E3A (UBE3A) also known as E6AP ubiquitin-protein ligase (E6AP) is an enzyme that in humans is encoded by the UBE3A gene. This enzyme is involved in targeting proteins for degradation within cells.
Protein degradation is a normal process that removes damaged or unnecessary proteins and helps maintain the normal functions of cells. Ubiquitin protein ligase E3A attaches a small marker protein called ubiquitin to proteins that should be degraded. Cellular structures called proteasomes recognize and digest proteins tagged with ubiquitin.
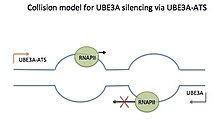
Both copies of the UBE3A gene are active in most of the body's tissues. In most neurons, however, only the copy inherited from a person's mother (the maternal copy) is normally active; this is known as paternal imprinting. Recent evidence shows that at least some glial cells and neurons may exhibit biallelic expression of UBE3A.[5][6] Further work is thus needed to delineate a complete map of UBE3A imprinting in humans and model organisms such as mice. Silencing of Ube3a on the paternal allele is thought to occur through the Ube3a-ATS part of a lincRNA called "LNCAT"[7] (Large Non-Coding Antisense Transcript).
The UBE3A gene is located on the long (q) arm of chromosome 15 between positions 11 and 13, from base pair 23,133,488 to base pair 23,235,220.
Clinical significance
Mutations within the UBE3A gene are responsible for some cases of Angelman syndrome and Prader-Willi syndrome. Most of these mutations result in an abnormally short, nonfunctional version of ubiquitin protein ligase E3A. Because the copy of the gene inherited from a person's father (the paternal copy) is normally inactive in the brain, a mutation in the remaining maternal copy prevents any of the enzyme from being produced in the brain. This loss of enzyme function likely causes the characteristic features of these two conditions.[citation needed]
The UBE3A gene lies within the human chromosomal region 15q11-13. Other abnormalities in this region of chromosome 15 can also cause Angelman syndrome. These chromosomal changes include deletions, rearrangements (translocations) of genetic material, and other abnormalities. Like mutations within the gene, these chromosomal changes prevent any functional ubiquitin protein ligase E3A from being produced in the brain.
UBE3A associates with the E6 protein of certain strains of HPV. This interaction promotes the polyubiquitination and subsequent degradation of the tumor suppressor gene p53, thereby enabling the immortalization of infected cells.[8] Strains of HPV with this ability have a higher risk of causing HPV-associated cancers. UBE3A is also known as E6AP or E6-associated protein in reference to this mechanism.
Interactions
UBE3A has been shown to interact with:
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000114062 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000025326 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Jones KA, Han JE, DeBruyne JP, Philpot BD (June 2016). "Persistent neuronal Ube3a expression in the suprachiasmatic nucleus of Angelman syndrome model mice". Scientific Reports. 6 (1): 28238. Bibcode:2016NatSR...628238J. doi:10.1038/srep28238. PMC 4910164. PMID 27306933.
- ^ Grier MD, Carson RP, Lagrange AH (2015-04-20). "Toward a Broader View of Ube3a in a Mouse Model of Angelman Syndrome: Expression in Brain, Spinal Cord, Sciatic Nerve and Glial Cells". PLOS ONE. 10 (4): e0124649. Bibcode:2015PLoSO..1024649G. doi:10.1371/journal.pone.0124649. PMC 4403805. PMID 25894543.
- ^ Runte M, Hüttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K (November 2001). "The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A". Human Molecular Genetics. 10 (23): 2687–700. doi:10.1093/hmg/10.23.2687. PMID 11726556.
- ^ Lehoux M, D'Abramo CM, Archambault J (2009). "Molecular Mechanisms of Human Papillomavirus-Induced Carcinogenesis". Public Health Genomics. 12 (5–6): 268–280. doi:10.1159/000214918. ISSN 1662-4246. PMC 4654617. PMID 19684440.
- ^ a b Oda H, Kumar S, Howley PM (August 1999). "Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination". Proceedings of the National Academy of Sciences of the United States of America. 96 (17): 9557–62. Bibcode:1999PNAS...96.9557O. doi:10.1073/pnas.96.17.9557. PMC 22247. PMID 10449731.
- ^ Kühne C, Banks L (December 1998). "E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box". The Journal of Biological Chemistry. 273 (51): 34302–9. doi:10.1074/jbc.273.51.34302. PMID 9852095.
- ^ Kim S, Chahrour M, Ben-Shachar S, Lim J (July 2013). "Ube3a/E6AP is involved in a subset of MeCP2 functions". Biochemical and Biophysical Research Communications. 437 (1): 67–73. doi:10.1016/j.bbrc.2013.06.036. PMID 23791832.
- ^ Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ, O'Malley BW (February 1999). "The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily". Molecular and Cellular Biology. 19 (2): 1182–9. doi:10.1128/mcb.19.2.1182. PMC 116047. PMID 9891052.
- ^ Lu Z, Hu X, Li Y, Zheng L, Zhou Y, Jiang H, Ning T, Basang Z, Zhang C, Ke Y (August 2004). "Human papillomavirus 16 E6 oncoprotein interferences with insulin signaling pathway by binding to tuberin". The Journal of Biological Chemistry. 279 (34): 35664–70. doi:10.1074/jbc.M403385200. PMID 15175323.
- ^ Zheng L, Ding H, Lu Z, Li Y, Pan Y, Ning T, Ke Y (March 2008). "E3 ubiquitin ligase E6AP-mediated TSC2 turnover in the presence and absence of HPV16 E6". Genes to Cells. 13 (3): 285–94. doi:10.1111/j.1365-2443.2008.01162.x. PMID 18298802.
- ^ a b Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M (February 1996). "Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5". The Journal of Biological Chemistry. 271 (5): 2795–800. doi:10.1074/jbc.271.5.2795. PMID 8576257.
- ^ Nuber U, Scheffner M (March 1999). "Identification of determinants in E2 ubiquitin-conjugating enzymes required for hect E3 ubiquitin-protein ligase interaction". The Journal of Biological Chemistry. 274 (11): 7576–82. doi:10.1074/jbc.274.11.7576. PMID 10066826.
- ^ a b Anan T, Nagata Y, Koga H, Honda Y, Yabuki N, Miyamoto C, Kuwano A, Matsuda I, Endo F, Saya H, Nakao M (November 1998). "Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes". Genes to Cells. 3 (11): 751–63. doi:10.1046/j.1365-2443.1998.00227.x. PMID 9990509. S2CID 1653536.
- ^ Hatakeyama S, Jensen JP, Weissman AM (June 1997). "Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases". The Journal of Biological Chemistry. 272 (24): 15085–92. doi:10.1074/jbc.272.24.15085. PMID 9182527.
- ^ Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (November 1999). "Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade". Science. 286 (5443): 1321–6. doi:10.1126/science.286.5443.1321. PMID 10558980.
- ^ a b Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM (August 2000). "The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome". Molecular Cell. 6 (2): 409–19. doi:10.1016/S1097-2765(00)00040-X. PMID 10983987.
Further reading
- Bittel DC, Kibiryeva N, Talebizadeh Z, Driscoll DJ, Butler MG (January 2005). "Microarray analysis of gene/transcript expression in Angelman syndrome: deletion versus UPD". Genomics. 85 (1): 85–91. doi:10.1016/j.ygeno.2004.10.010. PMC 6800218. PMID 15607424.
- Cassidy SB, Dykens E, Williams CA (2000). "Prader-Willi and Angelman syndromes: sister imprinted disorders". American Journal of Medical Genetics. 97 (2): 136–46. doi:10.1002/1096-8628(200022)97:2<136::AID-AJMG5>3.0.CO;2-V. PMID 11180221.
- Clayton-Smith J, Laan L (February 2003). "Angelman syndrome: a review of the clinical and genetic aspects". Journal of Medical Genetics. 40 (2): 87–95. doi:10.1136/jmg.40.2.87. PMC 1735357. PMID 12566516.
- Fang P, Lev-Lehman E, Tsai TF, Matsuura T, Benton CS, Sutcliffe JS, Christian SL, Kubota T, Halley DJ, Meijers-Heijboer H, Langlois S, Graham JM, Beuten J, Willems PJ, Ledbetter DH, Beaudet AL (January 1999). "The spectrum of mutations in UBE3A causing Angelman syndrome" (PDF). Human Molecular Genetics. 8 (1): 129–35. doi:10.1093/hmg/8.1.129. PMID 9887341.
- Moncla A, Malzac P, Livet MO, Voelckel MA, Mancini J, Delaroziere JC, Philip N, Mattei JF (July 1999). "Angelman syndrome resulting from UBE3A mutations in 14 patients from eight families: clinical manifestations and genetic counselling". Journal of Medical Genetics. 36 (7): 554–60. doi:10.1136/jmg.36.7.554. PMC 1734398. PMID 10424818.
- Williams CA (March 2005). "Neurological aspects of the Angelman syndrome". Brain & Development. 27 (2): 88–94. doi:10.1016/j.braindev.2003.09.014. PMID 15668046. S2CID 11172742.
External links
- GeneReviews/NCBI/NIH/UW entry on Angelman syndrome
- OMIM entries on Angelman syndrome
- GeneCard
- v
- t
- e
-
 1c4z: STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY
1c4z: STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY -
 1d5f: STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY
1d5f: STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY